Tidying the Johns Hopkins Covid-19 data
Update 2020-03-30: I have decided that the world needs another Covid19 related R package. Not sure whether you agree, but the new package facilitates the direct download of various Covid-19 related data (including data on governmental measures) directly from the authoritative sources. A short blog post about it can be found here. It also provides a flexible function and accompanying shiny app to visualize the spreading of the virus. Play around with the shiny app here if you like. The repo below stays in place for those of you that rather want to take data collection in their own hands. Please feel free to fork and explore. Stay well and keep #FlattenTheCurve!
Update 2020-03-28: I have created a github repo containing the updated code from below and more. I will try to maintain its code base moving forward. Please feel free to fork and explore. Stay well and keep #FlattenTheCurve!
Please Note: In the meantime, the Johns Hopkins CSSE team has adjusted the time-series table structure in their github repository. Currently, they only seem to provide time series data on confirmed cases and deaths. This new blog post reflects the changes. I am leaving the old blog in case somebody is interested in the data on recoveries.
My guess is that by now everybody knows that the public Github repository maintained by the Johns Hopkins University Center for Systems Science and Engineering has developed to a standard resource for individuals interested in analyzing the spread of SARS-CoV-2. There are alternative resources and blog articles covering them. Also, this blog post features a nice collection of R projects related to the Corona Virus.
The main reason for me sharing this code regardless is that I did not find code that merges standardized country level identifiers to to the data in a semi-automatic way. These identifiers are important whenever you want to merge additional country level data for additional analyses, like, e.g. population data to calculate per capita measures. Also, I thought that the steps presented below are nice small case on how to obtain, tidy and merge country-level data from public sources.
Pulling and tidying the Johns Hopkins Covid-19 data to long format
So, without much ado, here is the code for pulling and tidying the data.
library(tidyverse)
library(lubridate)
library(rvest)
library(stringdist)
# Function to read the raw CSV files. The files are aggregated to the country
# level and then converted to long format
clean_jhd_to_long <- function(df) {
df_str <- deparse(substitute(df))
var_str <- substr(df_str, 1, str_length(df_str) - 4)
df %>% group_by(`Country/Region`) %>%
filter(`Country/Region` != "Cruise Ship") %>%
select(-`Province/State`, -Lat, -Long) %>%
mutate_at(vars(-group_cols()), sum) %>%
distinct() %>%
ungroup() %>%
rename(country = `Country/Region`) %>%
pivot_longer(
-country,
names_to = "date_str",
values_to = var_str
) %>%
mutate(date = mdy(date_str)) %>%
select(country, date, !! sym(var_str))
}
confirmed_raw <- read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_19-covid-Confirmed.csv")
deaths_raw <- read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_19-covid-Deaths.csv")
recovered_raw <- read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_19-covid-Recovered.csv")
jh_covid19_data <- clean_jhd_to_long(confirmed_raw) %>%
full_join(clean_jhd_to_long(deaths_raw)) %>%
full_join(clean_jhd_to_long(recovered_raw))
# Next, I pull official country level indicators from the UN Statstics Division
# to get country level identifiers.
jhd_countries <- tibble(country = unique(jh_covid19_data$country)) %>% arrange(country)
ctry_ids <- read_html("https://unstats.un.org/unsd/methodology/m49/") %>%
html_table()
un_m49 <- ctry_ids[[1]]
colnames(un_m49) <- c("country", "un_m49", "iso3c")
# Merging by country name is messy. I start with a fuzzy matching approach
# using the {stringdist} package
ctry_names_dist <- matrix(NA, nrow = nrow(jhd_countries), ncol = nrow(un_m49))
for(i in 1:length(jhd_countries$country)) {
for(j in 1:length(un_m49$country)) {
ctry_names_dist[i,j]<-stringdist(tolower(jhd_countries$country[i]),
tolower(un_m49$country[j]))
}
}
min_ctry_name_dist <- apply(ctry_names_dist, 1, min)
matched_ctry_names <- NULL
for(i in 1:nrow(jhd_countries)) {
un_m49_row <- match(min_ctry_name_dist[i], ctry_names_dist[i,])
if (length(which(ctry_names_dist[i,] %in% min_ctry_name_dist[i])) > 1) un_m49_row <- NA
matched_ctry_names <- rbind(matched_ctry_names,
tibble(
jhd_countries_row = i,
un_m49_row = un_m49_row,
jhd_ctry_name = jhd_countries$country[i],
un_m49_name = ifelse(is.na(un_m49_row), NA,
un_m49$country[un_m49_row])
))
}
# This matches most cases well but some cases need to be adjusted by hand.
# In addition there are two jurisdictions (Kosovo, Taiwan)
# that cannot be matched as they are no 'country' as far as the U.N.
# Statistics Devision is concerned.
# WATCH OUT: The data from JHU is subject to change without notice.
# New countries are being added and names/spelling might change.
# Also, in the long run, the data provided by the UNSD might change.
# Inspect 'matched_ctry_names' before using the data.
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Bolivia"] <- 27
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Brunei"] <- 35
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Congo (Brazzaville)"] <- 54
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Congo (Kinshasa)"] <- 64
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "East Timor"] <- 222
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Iran"] <- 109
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Korea, South"] <- 180
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Kosovo"] <- NA
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Moldova"] <- 181
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Russia"] <- 184
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Taiwan*"] <- NA
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Tanzania"] <- 236
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "United Kingdom"] <- 235
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "US"] <- 238
matched_ctry_names$un_m49_row[matched_ctry_names$jhd_ctry_name == "Venezuela"] <- 243
# Last Step: Match country identifier data and save file (commented out here)
jhd_countries %>%
left_join(matched_ctry_names %>%
select(jhd_ctry_name, un_m49_row),
by = c(country = "jhd_ctry_name")) %>%
left_join(un_m49 %>% mutate(un_m49_row = row_number()), by = "un_m49_row") %>%
rename(country = country.x) %>%
select(country, iso3c) -> jhd_countries
jh_covid19_data <- jh_covid19_data %>% left_join(jhd_countries) %>%
select(country, iso3c, date, confirmed, deaths, recovered)
# write_csv(jh_covid19_data, sprintf("jh_covid19_data_%s.csv", Sys.Date()))The code essentially follows the following steps
- Read the relevant CSV files for confirmed cases, casualties and recovered patients from the Github repository
- Aggregate the data at country level and discard data that is not required
- Scrape official country identifiers from the U.N. Statistics Division
- Fuzzy match these to the countries present in the JH data. Apply manual corrections that were correct based on the data pulled March 23, 2020 (check the match when you use this code later)
- Merge the identifiers with the longitudinal data and save the result as a tidy CSV file.
Merging some World Bank data
The next code snippet pulls some World Bank data using the {wbstats} package.
library(tidyverse)
library(wbstats)
pull_worldbank_data <- function(vars) {
new_cache <- wbcache()
all_vars <- as.character(unique(new_cache$indicators$indicatorID))
data_wide <- wb(indicator = vars, mrv = 10, return_wide = TRUE)
new_cache$indicators[new_cache$indicators[,"indicatorID"] %in% vars, ] %>%
rename(var_name = indicatorID) %>%
mutate(var_def = paste(indicator, "\nNote:",
indicatorDesc, "\nSource:", sourceOrg)) %>%
select(var_name, var_def) -> wb_data_def
new_cache$countries %>%
select(iso3c, iso2c, country, region, income) -> ctries
left_join(data_wide, ctries, by = "iso3c") %>%
rename(year = date,
iso2c = iso2c.y,
country = country.y) %>%
select(iso3c, iso2c, country, region, income, everything()) %>%
select(-iso2c.x, -country.x) %>%
filter(!is.na(NY.GDP.PCAP.KD),
region != "Aggregates") -> wb_data
wb_data$year <- as.numeric(wb_data$year)
wb_data_def<- left_join(data.frame(var_name = names(wb_data),
stringsAsFactors = FALSE),
wb_data_def, by = "var_name")
wb_data_def$var_def[1:6] <- c(
"Three letter ISO country code as used by World Bank",
"Two letter ISO country code as used by World Bank",
"Country name as used by World Bank",
"World Bank regional country classification",
"World Bank income group classification",
"Calendar year of observation"
)
wb_data_def$type = c("cs_id", rep("factor", 4), "ts_id",
rep("numeric", ncol(wb_data) - 6))
return(list(wb_data, wb_data_def))
}
vars <- c("SP.POP.TOTL", "AG.LND.TOTL.K2", "EN.POP.DNST", "EN.URB.LCTY", "SP.DYN.LE00.IN", "NY.GDP.PCAP.KD")
wb_list <- pull_worldbank_data(vars)
wb_data <- wb_list[[1]]
wb_data_def <- wb_list[[2]]
wb_data %>%
group_by(iso3c) %>%
arrange(iso3c, year) %>%
summarise(
population = last(na.omit(SP.POP.TOTL)),
land_area_skm = last(na.omit(AG.LND.TOTL.K2)),
pop_density = last(na.omit(EN.POP.DNST)),
pop_largest_city = last(na.omit(EN.URB.LCTY)),
gdp_capita = last(na.omit(NY.GDP.PCAP.KD)),
life_expectancy = last(na.omit(SP.DYN.LE00.IN))
) %>% left_join(wb_data %>% select(iso3c, region, income) %>% distinct()) -> wb_cs
# write_csv(wb_cs, "jh_add_wbank_data.csv")Use the data
And finally, some code to use the data for typical event time visualizations.
suppressPackageStartupMessages({
library(tidyverse)
library(lubridate)
library(gghighlight)
library(ggrepel)
})## Warning: package 'ggplot2' was built under R version 3.6.2## Warning: package 'tibble' was built under R version 3.6.2## Warning: package 'tidyr' was built under R version 3.6.2## Warning: package 'readr' was built under R version 3.6.2## Warning: package 'purrr' was built under R version 3.6.2## Warning: package 'dplyr' was built under R version 3.6.2## Warning: package 'lubridate' was built under R version 3.6.2dta <- read_csv(
"https://joachim-gassen.github.io/data/jh_covid19_data_2020-03-23.csv",
col_types = cols()
) %>%
mutate(date = ymd(date))
wb_cs <- read_csv(
"https://joachim-gassen.github.io/data/jh_add_wbank_data.csv",
col_types = cols()
)
# I define event time zero where, for a given country, the confirmed
# cases match or exceed the Chinese case number at the beginning of the
# data so that all countries can be compared across event time.
# Also a require each country to have at least 7 days post event day 0
dta %>%
group_by(country) %>%
filter(confirmed >= min(dta$confirmed[dta$country == "China"])) %>%
summarise(edate_confirmed = min(date)) -> edates_confirmed## `summarise()` ungrouping output (override with `.groups` argument)dta %>%
left_join(edates_confirmed, by = "country") %>%
mutate(
edate_confirmed = as.numeric(date - edate_confirmed)
) %>%
filter(edate_confirmed >= 0) %>%
group_by(country) %>%
filter (n() >= 7) %>%
ungroup() %>%
left_join(wb_cs, by = "iso3c") %>%
mutate(
confirmed_1e5pop = 1e5*confirmed/population
) -> df
lab_notes <- paste0(
"Data as provided by Johns Hopkins University Center for Systems Science ",
"and Engineering (JHU CSSE)\nand obtained on March 23, 2020. ",
"The sample is limited to countries with at least seven days of positive\n",
"event days data. Code and walk-through: https://joachim-gassen.github.io."
)
lab_x_axis_confirmed <- sprintf(paste(
"Days since confirmed cases matched or exceeded\n",
"initial value reported for China (%d cases)\n"
), min(dta$confirmed[dta$country == "China"]))
gg_my_blob <- list(
scale_y_continuous(trans='log10', labels = scales::comma),
theme_minimal(),
theme(
plot.title.position = "plot",
plot.caption.position = "plot",
plot.caption = element_text(hjust = 0),
axis.title.x = element_text(hjust = 1),
axis.title.y = element_text(hjust = 1),
),
labs(caption = lab_notes,
x = lab_x_axis_confirmed,
y = "Confirmed cases (logarithmic scale)"),
gghighlight(TRUE, label_key = country, use_direct_label = TRUE,
label_params = list(segment.color = NA, nudge_x = 1))
)
ggplot(df %>% filter (edate_confirmed <= 30),
aes(x = edate_confirmed, color = country, y = confirmed)) +
geom_line() +
labs(
title = "Focus on the first month: Confirmed Cases\n"
) +
gg_my_blob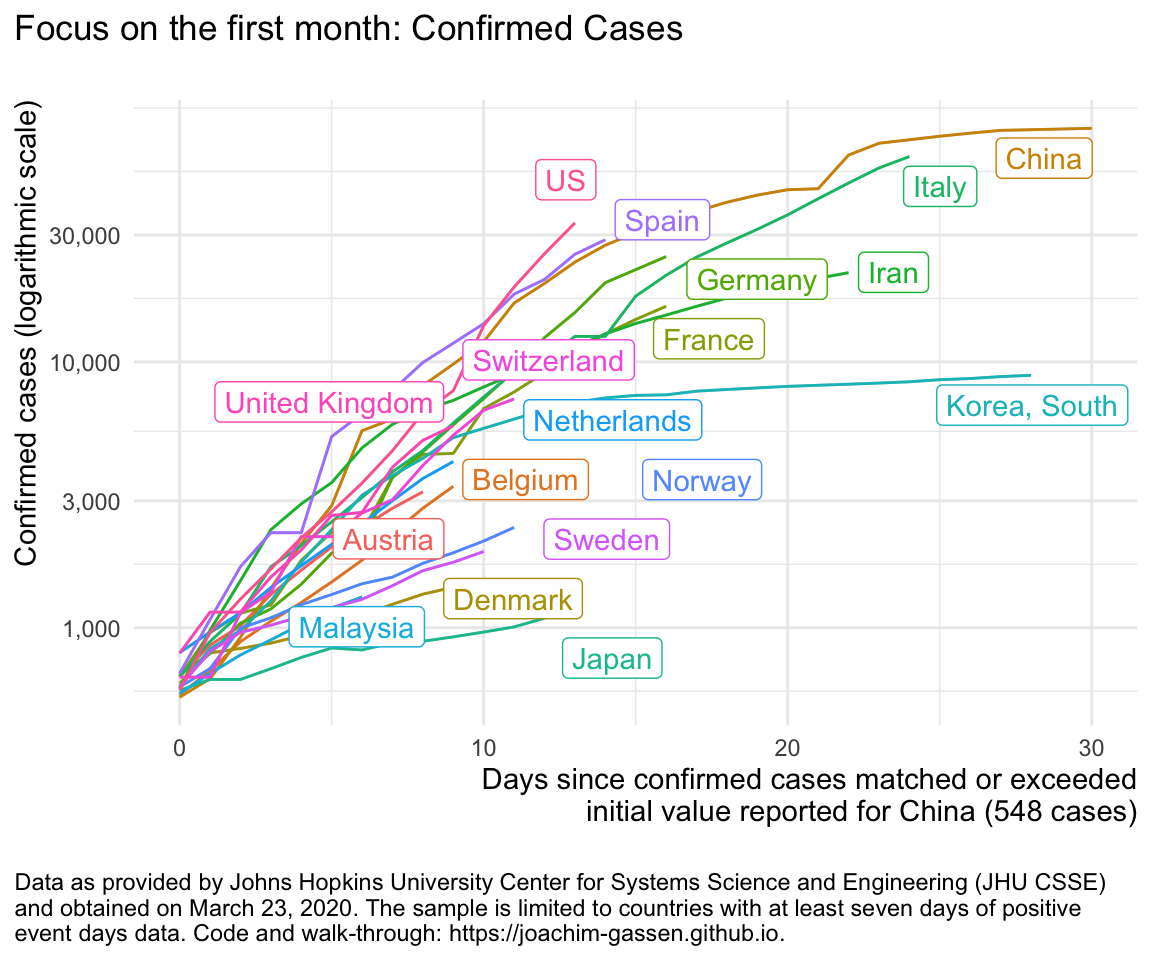
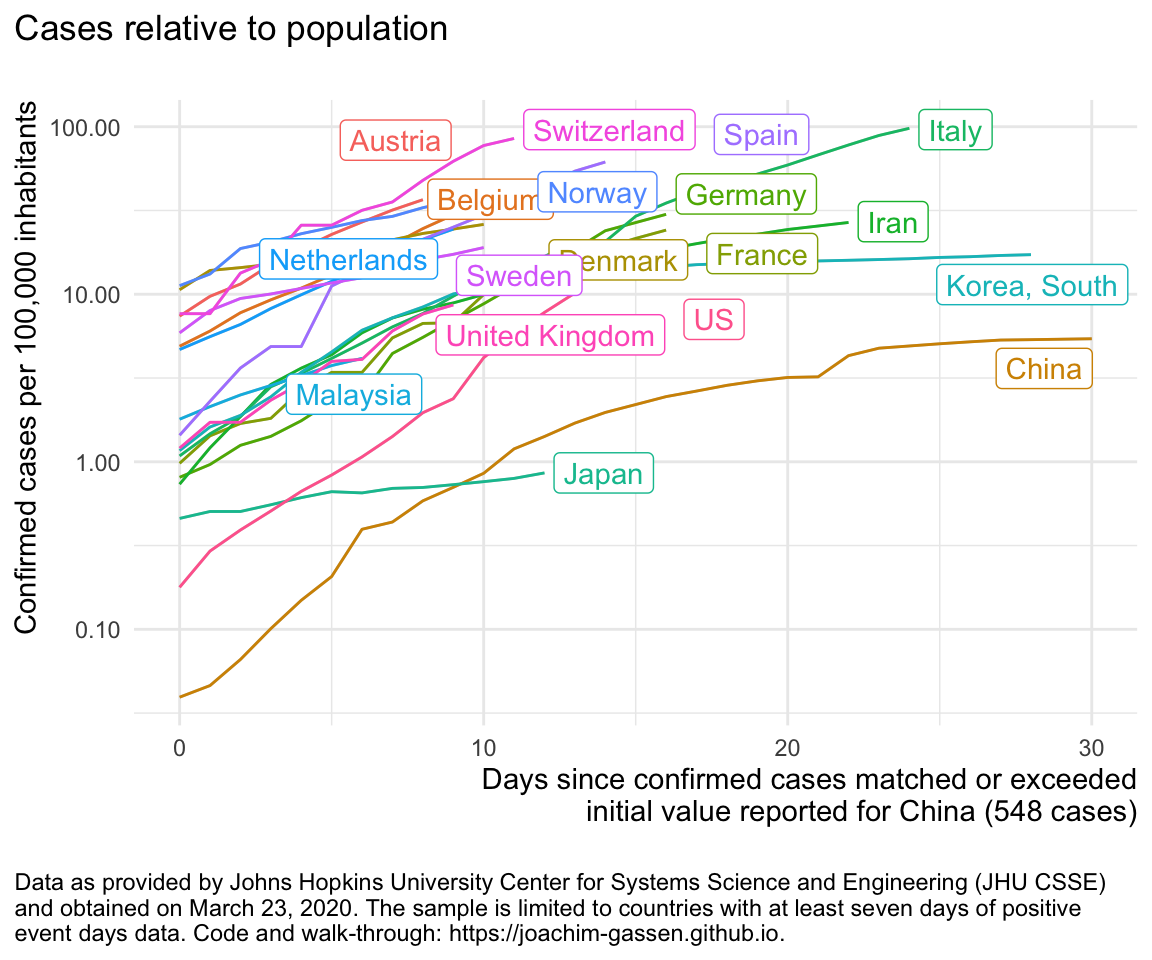
ggplot(df %>% filter (edate_confirmed <= 30),
aes(x = edate_confirmed, color = country, y = confirmed_1e5pop)) +
geom_line() +
gg_my_blob +
labs(
y = "Confirmed cases per 100,000 inhabitants",
title = "Cases relative to population\n"
) 
Wrap-Up
This is it. I hope that somebody might fight this useful. In any case, help #FlattenTheCurve and stay healthy, everybody!
