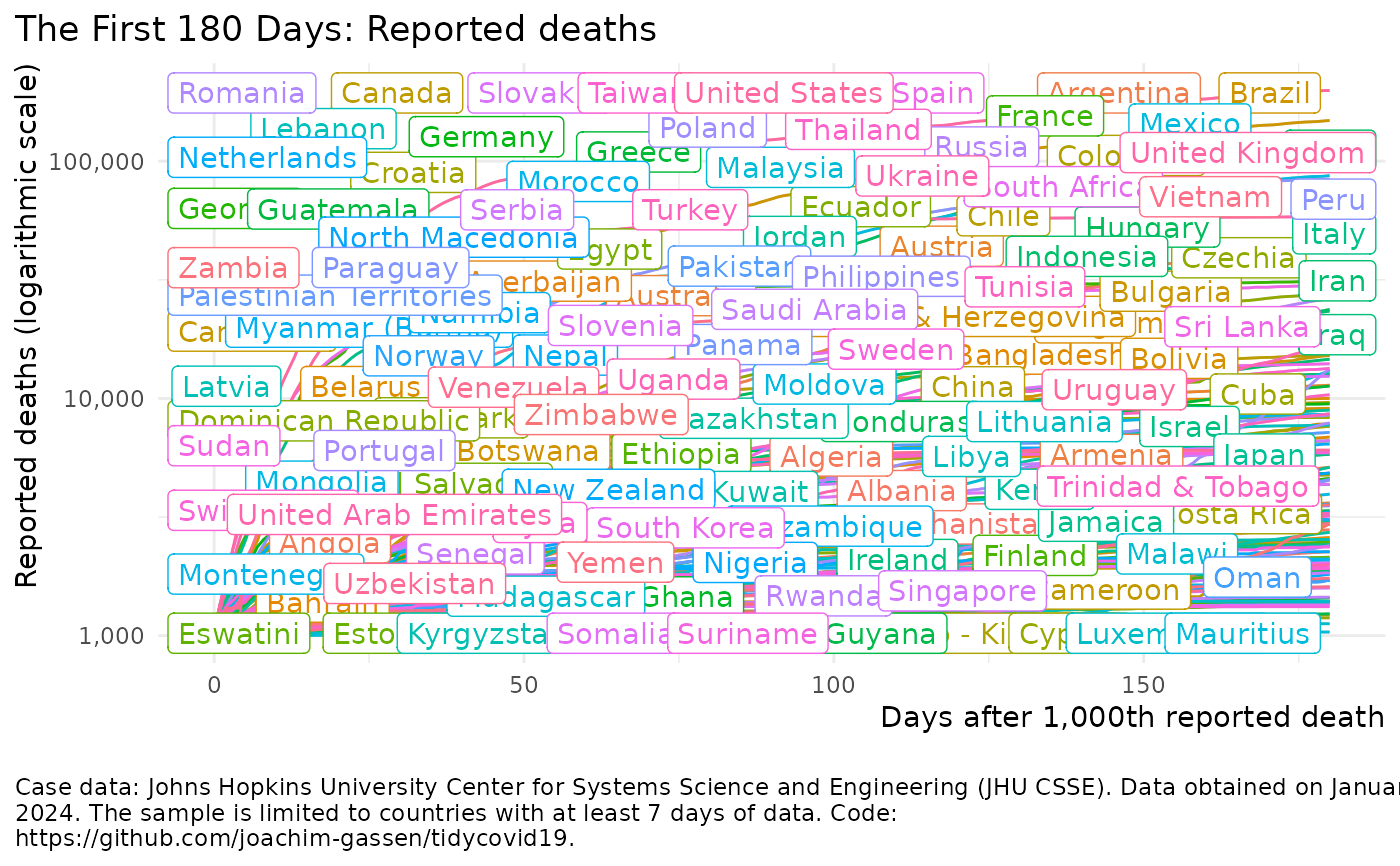
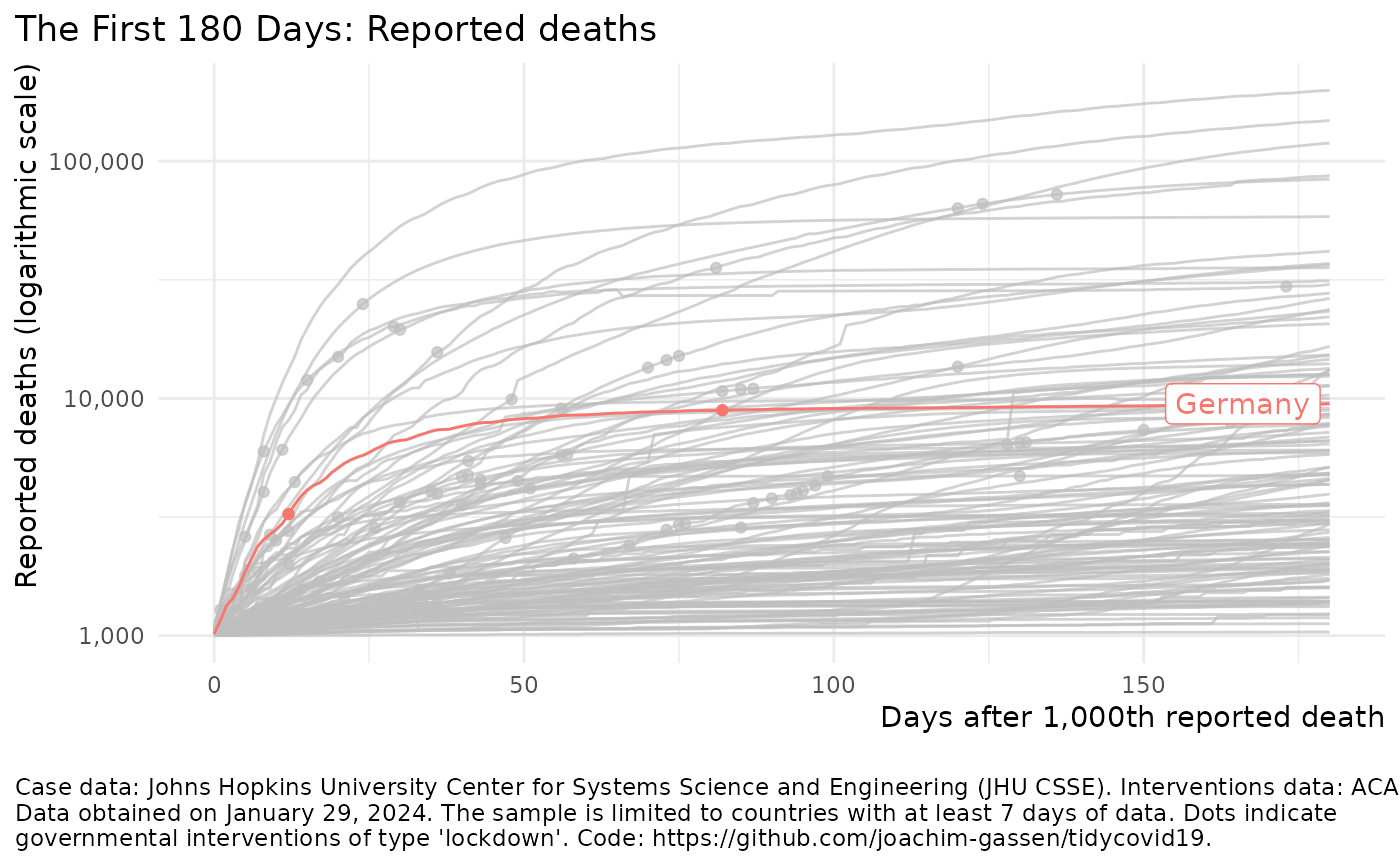
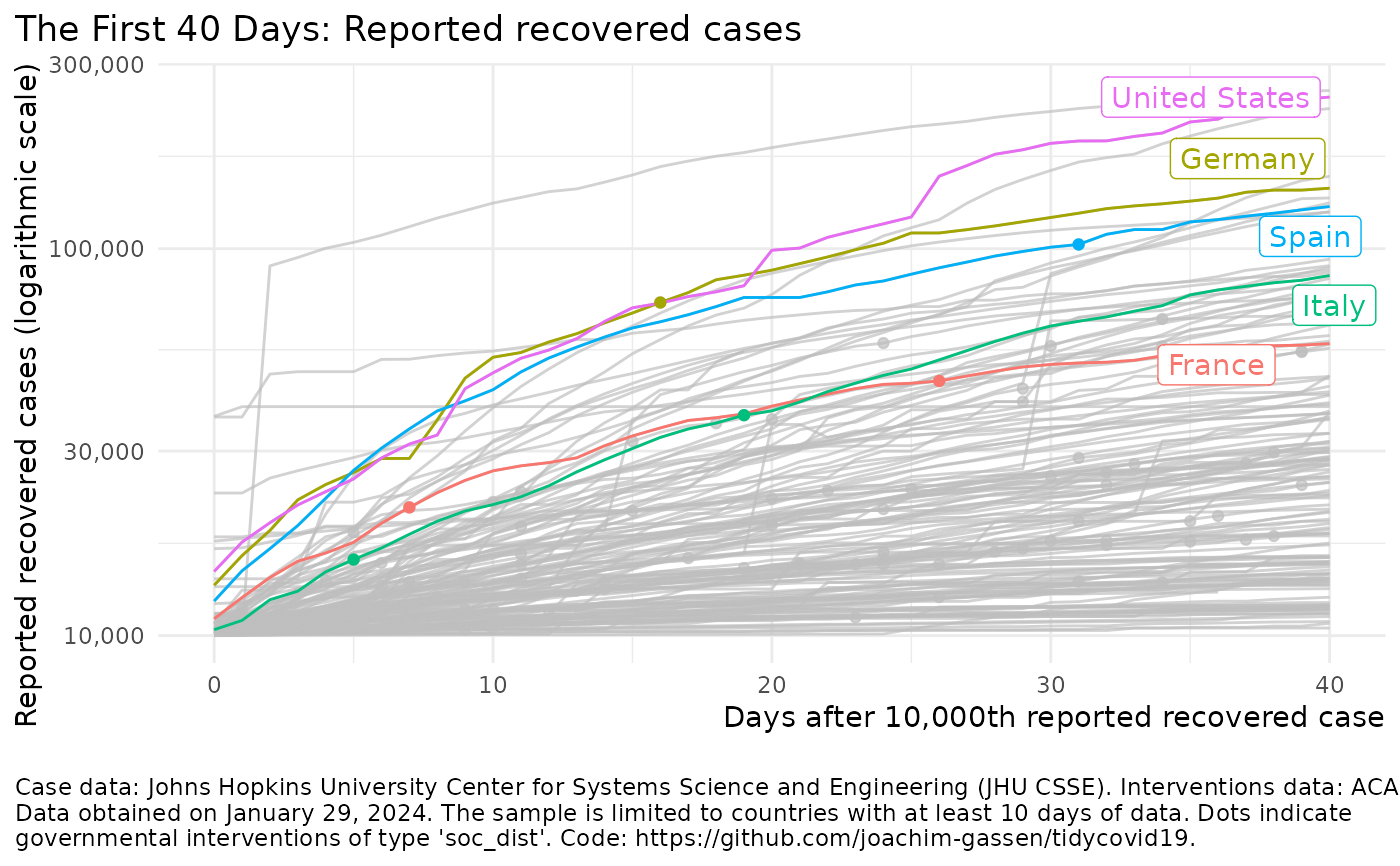
Visualize the spread of the Covid-19 pandemic
plot_covid19_spread.RdProvides a flexible visualization of the country-level Covid-19 spread,
inpired by the displays created by John Burn-Murdoch from the Financial
Times. Uses data from the Johns Hopkins University CSSE team
(https://github.com/CSSEGISandData/COVID-19), the ACAPS governmental
measures database
(https://www.acaps.org/covid19-government-measures-dataset), and the
World Bank (https://data.worldbank.org).
If your are overwhelmed with the options of the plot, explore them with
shiny_covid19_spread() and use the 'copy code for plot to clipboard'
option.
plot_covid19_spread( data = download_merged_data(cached = TRUE, silent = TRUE), type = "deaths", min_cases = ifelse(per_capita, ifelse(type == "deaths", 5, 50), ifelse(type == "deaths", 1000, 10000)), min_by_ctry_obs = 7, edate_cutoff = 180, data_date_str = format(lubridate::as_date(data$timestamp[1]), "%B %d, %Y"), cumulative = TRUE, change_ave = 7, per_capita = FALSE, per_capita_x_axis = ifelse(per_capita, TRUE, FALSE), population_cutoff = 0, log_scale = TRUE, highlight = NULL, exclude_others = FALSE, intervention = NULL )
Arguments
| data | The data frame to base the plot on. Should be a merged data
frame obtained by download_merged_data and defaults to
|
|---|---|
| type | The statistic that you want to plot. Needs to be either "confirmed", "deaths", "recovered" or "active", defined as the difference of "confirmed", "deaths" and "recovered". |
| min_cases | Defines the zero point of your X axis (the 'event date'). Defaults to 1,000 cases for deaths and 10,000 cases otherwise. |
| min_by_ctry_obs | Limits the plot to countries that have at least that many days of data since and including the event date. Defaults to 7. |
| edate_cutoff | The upper limit of the X axis in event days. Defaults to 180. |
| data_date_str | A date string to include in the annotation of the plot giving the time when the data was pulled. Defaults to the timestamp of the data. Note that you might run into issues with the default when running this in a non-english locale. Consider setting it by hand then. |
| cumulative | If |
| change_ave | Number of days to average over when you plot daily changes. |
| per_capita | If |
| per_capita_x_axis | If |
| population_cutoff | Do you want to restrict the plot to countries that exceed a certain population cutoff? Takes a value in millions and defaults to 0. Useful for per capita displays. |
| log_scale | Do you want the Y-axis to be log-scaled? Defaults to
|
| highlight | A character vector of ISO3c (ISO 3166-1 alpha-3) codes that
identify countries that you want to highlight. Using the
|
| exclude_others | If |
| intervention | If not default |
Value
A ggplot2 object.
Examples
plot_covid19_spread()#> Failed to get face: , 22041merged <- download_merged_data(cached = TRUE, silent = TRUE) plot_covid19_spread(merged, highlight = "DEU", intervention = "lockdown")plot_covid19_spread(merged, type = "recovered", min_by_ctry_obs = 10, edate_cutoff = 40, highlight = c("ITA", "ESP", "FRA", "DEU", "USA"), intervention = "soc_dist" )