Download a merged country-day data frame containing Covid-19 related data
download_merged_data.RdMerges data from the Johns Hopkins University CSSE team on the spread of the
SARS-CoV-2 virus and the Covid-19 pandemic
(https://github.com/CSSEGISandData/COVID-19),
case data provided by the ECDC
(https://www.ecdc.europa.eu/en/publications-data/download-todays-data-geographic-distribution-covid-19-cases-worldwide),
the ACAPS governmental measures database
(https://www.acaps.org/covid19-government-measures-dataset),
the Oxford Covid-19 Government Respoonse Tracker
(https://github.com/OxCGRT/covid-policy-tracker),
Mobility Trends Reports provided by Apple related to Covid-19
(https://www.apple.com/covid19/mobility),
Google COVID-19 Community Mobility Reports
(https://www.google.com/covid19/mobility/),
Google Trends Covid-19 related search volume
(https://trends.google.com/trends/),
data on tests, vaccinations and hospitalizations as collected by the
Our World in Data team,
(https://github.com/owid/covid-19-data/tree/master/public/data),
and from the World Bank
(https://data.worldbank.org) intro a country-day data frame.
Variable definitions are provided by the data frame
tidycovid19_variable_definitions that also reports on the current
status of the data.
download_merged_data( wbank_vars = c("SP.POP.TOTL", "AG.LND.TOTL.K2", "EN.POP.DNST", "EN.URB.LCTY", "SP.DYN.LE00.IN", "NY.GDP.PCAP.KD"), wbank_labels = c("population", "land_area_skm", "pop_density", "pop_largest_city", "life_expectancy", "gdp_capita"), search_term = "coronavirus", silent = FALSE, cached = FALSE )
Arguments
| wbank_vars | Specify the World Bank data items that you want to retrieve. |
|---|---|
| wbank_labels | Give somewhat more informative World Bank variable names
for the output data frame. Has to match the length of |
| search_term | Google Trends serch term. Defaults to "coronavirus". |
| silent | Whether you want the function to send some status messages to
the console. Might be informative as downloading will take some time
and thus defaults to |
| cached | Whether you want to download the cached version of the data
from the tidycovid19 Github repository instead of retrieving the
data from the authorative source. Downloading the cached version is
faster and the cache is updated daily. Defaults to |
Value
A data frame containing the data, organized by country and date. It
includes a timestamp variable indicating the time of data
retrieval.
Details
See the documentation of the separate download functions of the package for more detail.
Examples
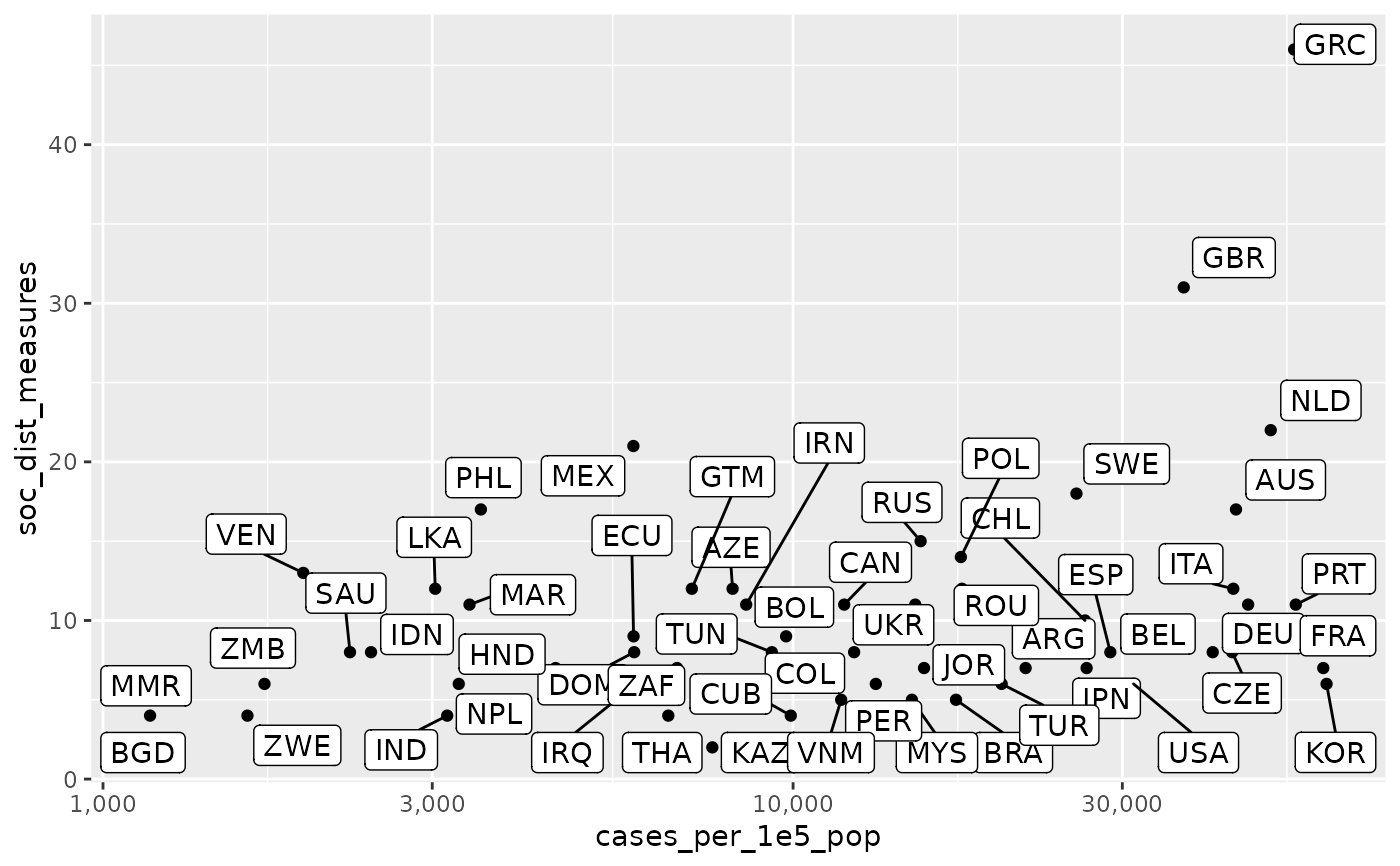
df <- download_merged_data(silent = TRUE, cached = TRUE) df %>% dplyr::group_by(iso3c) %>% dplyr::filter(population > 10e6) %>% dplyr::summarise( cases_per_1e5_pop = max(1e5*(confirmed/population), na.rm = TRUE), soc_dist_measures = max(soc_dist, na.rm = TRUE), .groups = "drop" ) %>% dplyr::filter(cases_per_1e5_pop >= 1000) %>% ggplot2::ggplot(mapping = ggplot2::aes(x = cases_per_1e5_pop, y = soc_dist_measures)) + ggplot2::geom_point() + ggrepel::geom_label_repel(ggplot2::aes(label = iso3c)) + ggplot2::scale_x_continuous(trans='log10', labels = scales::comma)